What is CRISPR?
Clustered regularly interspaced short palindromic repeats (CRISPR) are sequences found in many bacteria and archaea which constitute one of the most effective prokaryotic defense systems against potential infectious agents including bacteriophages and viruses. A plethora of CRISPR-Cas related studies not only broadened our understanding of the interactions between prokaryotes and their phages and viruses, but they also gave researchers the opportunity to apply CRISPR-based machinery in genetic modification experiments. Refer to this blog post if you want to learn more about the history of CRISPR and CRISPR-Cas. The continuous development of novel CRISPR-based genome editing tools consistently leads to major advances in the life sciences [1]. This is one of the reasons why it is important to carefully follow up-to-date tips and tricks on how to maximize the efficiency and precision of your CRISPR experiments.
CRISPR knock-ins vs. knock-outs
To implement targeted genome edits, researchers can use two main approaches: knock-ins or knock-outs [2]. Before discussing the actual tips for a successful knock-in experiment, let’s briefly overview what the CRISPR system consists of.
The CRISPR-based machinery involves two main components: (i) a guide RNA, a specific RNA sequence that recognizes the targeted DNA region and (ii) a Cas nuclease, most commonly Cas9, which is directed to a specific DNA site by the RNA guide and cuts at
this DNA site (Figure 1). Sometimes these cuts result in double-strand breaks (DSBs) in the chromosome which may lead to cell death. To avoid cell death, DSBs can be repaired by one of the following mechanisms: microhomology-mediated end joining (MMEJ),
non-homologous end joining (NHEJ), or homology-directed repair (HDR).
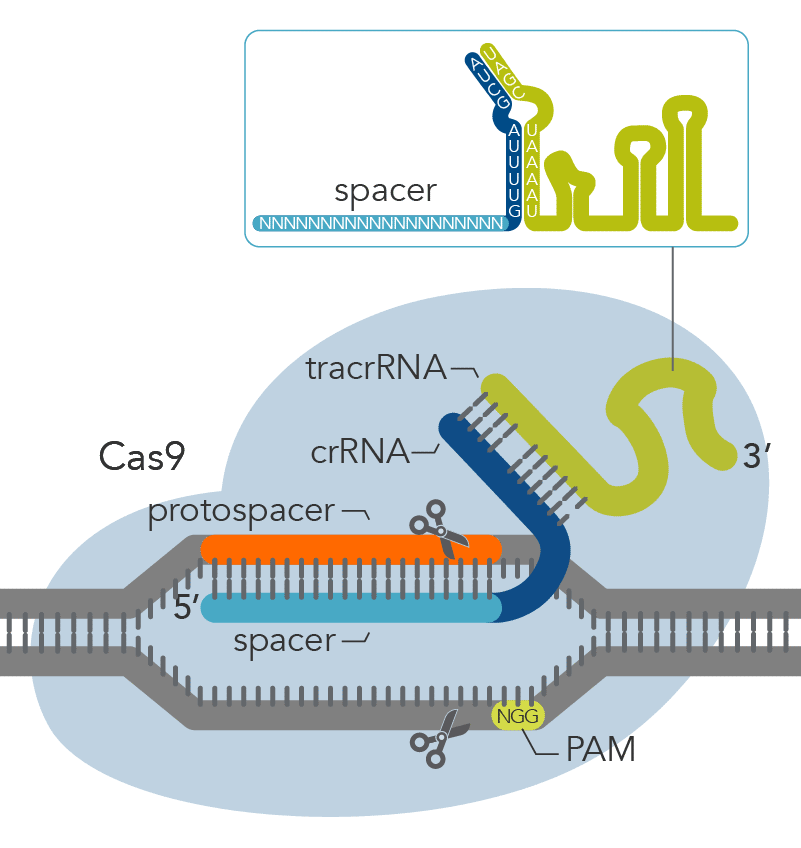
Figure 1. Components of the Alt-R™ CRISPR-Cas9 System for directing Cas9 endonuclease to genomic targets.
NHEJ, often used for knock-out experiments, repairs the DSB rapidly, often creating indels (insertions or deletions) which may lead to a frameshift mutation. On the other hand, knock-in experiments—the main goal of which is to insert an exact DNA sequence into a precise genomic site (which leaves virtually no room for error)—rely on the HDR mechanism. Since HDR-mediated repair is only active in dividing cells and given the high precision of the necessary edit, knock-in experiments tend to be more challenging compared to knock-out experiments.
Tips for a successful knock-in experiment
What steps can be taken to maximize the efficiency and precision of your CRISPR knock-in experiment?
- Choose the right guide RNA
The guide RNA plays a key role in a knock-in experiment. It targets a highly specific DNA sequence and guides the Cas nuclease to this site. This is why it is important to carefully design your guide RNA using one of the freely available bioinformatics tools, for example, IDT’s Alt‑R HDR Design Tool. It is also recommended to design a few different guide RNAs and assess their efficiency to achieve the best results. - Pick the right DNA donor template
The success of your knock-in experiment also relies on choosing the right type of the DNA donor template. This choice largely depends on the size of your insertion. For short insertions (<120 bp), it is recommended to use single-stranded donor oligonucleotides (ssODN). Whereas for longer insertions, it is advisable to use a plasmid or double-stranded DNA (dsDNA) due to synthesis limits of oligonucleotides. It is worth noting that using plasmids may result in lower knock-in efficiency because large plasmid constructs are difficult to deliver efficiently and may cause toxicity. IDT recommends the use of chemical modifications to stabilize your donor template against nucleases and reduce the risk of non-HDR mediated blunt insertions. Our Alt‑R HDR Donor Oligos and Alt‑R HDR Donor Blocks include these modifications. - Consider silent mutations
Adding silent mutations into the protospacer adjacent motif (PAM) or the target site may have a double positive effect of preventing premature degradation of the dsDNA template and preventing recutting of the genomic target DNA. If the photospacer is disrupted by the introduction of a silent mutation, the gRNA-Cas9 complex can’t bind to the site and the nuclease can’t re-cut the target locus. It is worth noting that adding silent mutations is a feature of the Alt‑R HDR Design Tool. For additional comprehensive design considerations for efficient HDR, refer to this article by Schubert et al., 2021 [3]. - Optimize experimental conditions
No matter what scientific experiment you are running, its conditions should be carefully adjusted to deliver the best results. This is especially important in the case of a CRISPR knock-in experiment that intrinsically has low efficiency. What experimental adjustments can be made for better results?- Choose the appropriate delivery system (e.g., electroporation) and delivery conditions
- Determine the optimal concentration of the gRNA-Cas9 complex and HDR template
- Experiment with initial cell density
- Adjust the guide RNA to Cas9 ratio if needed (typically, around 1.2:1)
- Incorporate electroporation and HDR enhancers to improve delivery and increase HDR rates
CRISPR products offered by IDT
The insertion (knock-in) of an additional DNA sequence into a genome is widely used for modifying genomes in experimental systems across multiple industries. Since the efficiency of knock-ins is naturally low, it is important to maximize the efficiency of your results by carefully following all the tips and tricks available thanks to the continual advancement of scientific knowledge and improved quality of CRISPR-related tools and reagents. For more information about CRISPR technology and the products offered by IDT for your experimental needs, you can download our free CRISPR Basics Handbook and read this application note about HDR.

