What is base editing?
Base editing is a genome editing method that aims to change a single base within a target sequence. Base editors have been developed to target either DNA or RNA. Of those designed to edit DNA, there are two main types: the cytosine base editor (CBE), which changes a cytosine-guanine pairing to a thymine‑adenine pairing [1] and the adenine base editor (ABE) which changes an adenine‑thymine pairing to a guanine-cytosine pairing [2].
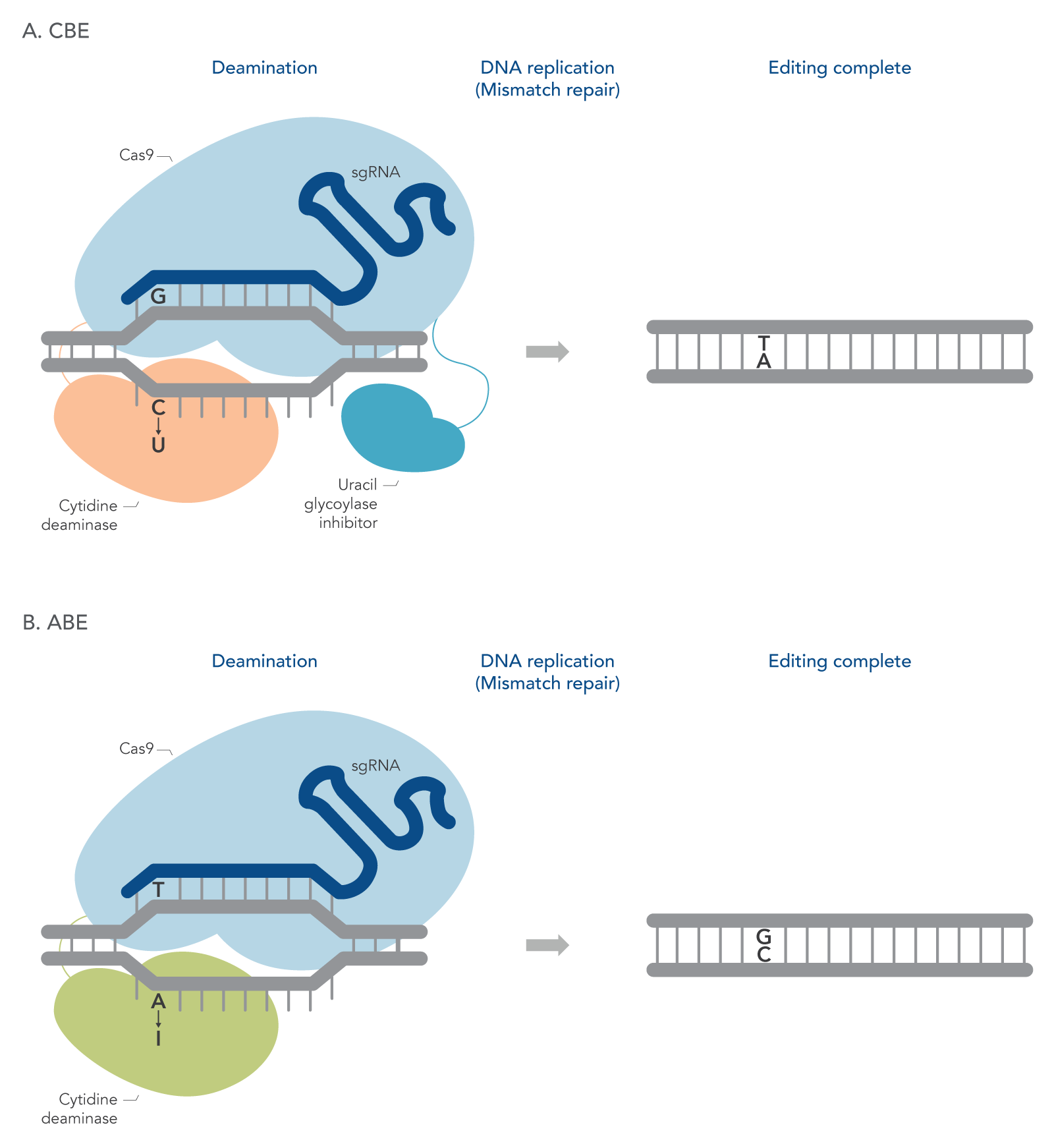
Figure 1. Simplified illustration of CBE and ABEs. The deaminase for each editor is linked to a Cas9 protein, which pairs with a sgRNA. The deaminase binds to a single strand of DNA and converts either cytosine to uridine [A(CBE)] or adenosine to inosine [B(ABE)], and during DNA replication the mismatch repair or base excision repair mechanism completes the edit by correcting the opposite strand to contain the appropriate nucleotide–thymine for the CBE and guanine for the ABE. The CBE also involves an uracil glycosylase inhibitor domain to ensure that the C-U edit is maintained during repair.
How does base editing work?
Base editing involves the targeted recruitment of a deaminase to convert one base to another without generating a double-stranded break. This can be achieved by using a single-stranded DNA deaminase domain, linked to a catalytically deficient CRISPR protein such as Cas9 (dCas9) or nickase, to modify bases in the free strand of DNA that is formed when the CRISPR protein binds to the targeted site within the genome (Figure 1). Including a nickase can improve editing efficiency as this enzyme will ‘nick’ the unmodified DNA strand, stimulating the DNA repair mechanism to use the modified strand as the template, thereby ensuring that the editing is complete.
For CBEs, deamination of cytosine forms uridine which is converted to thymine through the natural mismatch repair pathways of the cell (Figure 1A). CBEs can also include an uracil glycosylase inhibitor domain to prevent the cells from detecting the U mismatch in the DNA and changing it back to a C (Figure 1A). For ABEs, recruitment of an adenosine deaminase converts adenosine to inosine which can then be paired with guanosine (Figure 1B).
CBEs and ABEs were initially generated in the lab of David Liu, PhD [1,2], and since then improvements upon the first generation of these editors have resulted in an increased editing efficiency, reduced off-target edits, and greater flexibility in genetic targets [3,4]. More recently, base editors that can perform both cytosine and adenine base editing (ACBE) have also been developed [5-7].
Are base editing and CRISPR the same?
Even though DNA base editing and CRISPR can both use some variation of a Cas9 protein to edit genes, there are key differences between the two approaches which are outlined in Table 1.
| Base editing | CRISPR/Cas9 |
|---|---|
| Efficient/fewer errors but limited in types of changes that can be made | Efficient for disrupting genes or knock‑in/knock‑out of large segments of DNA |
| Needs PAM site close to target, multiple As or Cs in a row can be difficult to limit editing effects | Larger flexibility around targets |
| Relatively new, off target effects not well understood | Well-established, larger understanding of off target effects |
| Relies on mismatch repair for edits | Double-stranded break needed for gene editing |
| No donor template needed | Need to supply a donor template for specific edit |
| Can efficiently be conducted in non-dividing cells | HDR only efficient in dividing cells |
More generally, gene editing with CRISPR results in a double-stranded break in the DNA and relies on non-homologous end joining (NHEJ) or homology-directed repair (HDR) to knock‑in or knock‑out large segments of DNA. Base editors take advantage of the base excision repair or mismatch repair mechanisms within a cell to change individual bases, without generating a double‑stranded break in the DNA [8]. Prime editing, another CRISPR‑adjacent gene editing technique developed by the Liu lab, also makes targeted edits without a double‑stranded break but uses a relatively lengthy guide RNA (pegRNA) to dictate the specified genetic alterations [9].
To find out more about the fundamentals of CRISPR-based genome editing, download The CRISPR Basics Handbook. You can also read more about prime editing here.
How can base editors be used?
Researchers have been using base editing to advance the field of genomic medicine by targeting single nucleotide variants (SNVs) which have been linked to pathogenic phenotypes. One approach is to use base editors to correct the problematic SNV, which has been applied by researchers studying genes associated with inherited diseases such as Marfan syndrome, cancer, and Duchenne muscular dystrophy [8]. Other ways researchers have been applying base editing include changing codons to either start or stop codons to disrupt transcription of problematic genes [8] or by inactivating splice-site motifs [10], as well as identifying SNVs in plants that confer positive functional traits in crops [11]. Even though initially, base editors were limited to making transition mutations (converting a purine base to different purine [e.g. A > G] or a pyrimidine to another pyrimidine [e.g., C > T]), researchers have continued to refine and expand the capabilities of this technology, thus opening up new potential applications and genetic targets [12,13].

