Background
Next generation sequencing (NGS) has found wide applicability, including determining and characterizing unknown sequences, detecting changes and variability in known sequences, and quantifying gene expression. One of the greatest advantages of NGS is the ability to sequence and analyze large numbers of samples simultaneously.
During library preparation, the DNA extracted from each sample is first fragmented, either mechanically or enzymatically, and then ligated with known index sequences. Several sample libraries, each tagged with unique index sequences, can then be pooled or multiplexed for sequencing [1].
Yet, sample processing on a high-throughput scale presents cross-contamination and sample swapping risks that result in data analysis errors and misassigned reads [2,3]. The use of plasmids and synthetic oligonucleotide fragments is recommended as spike-in controls for NGS applications to identify sample swaps and cross-contamination, as well as measure various assay parameters [2–5].
gBlocks Gene Fragments
gBlocks Gene Fragments are linear, double-stranded DNA
fragments that can be custom-designed as spike-in controls for NGS. Adding known controls that are subject to the same experimental conditions as the sample can be used to quantify technical bias and track samples (Figure 1). 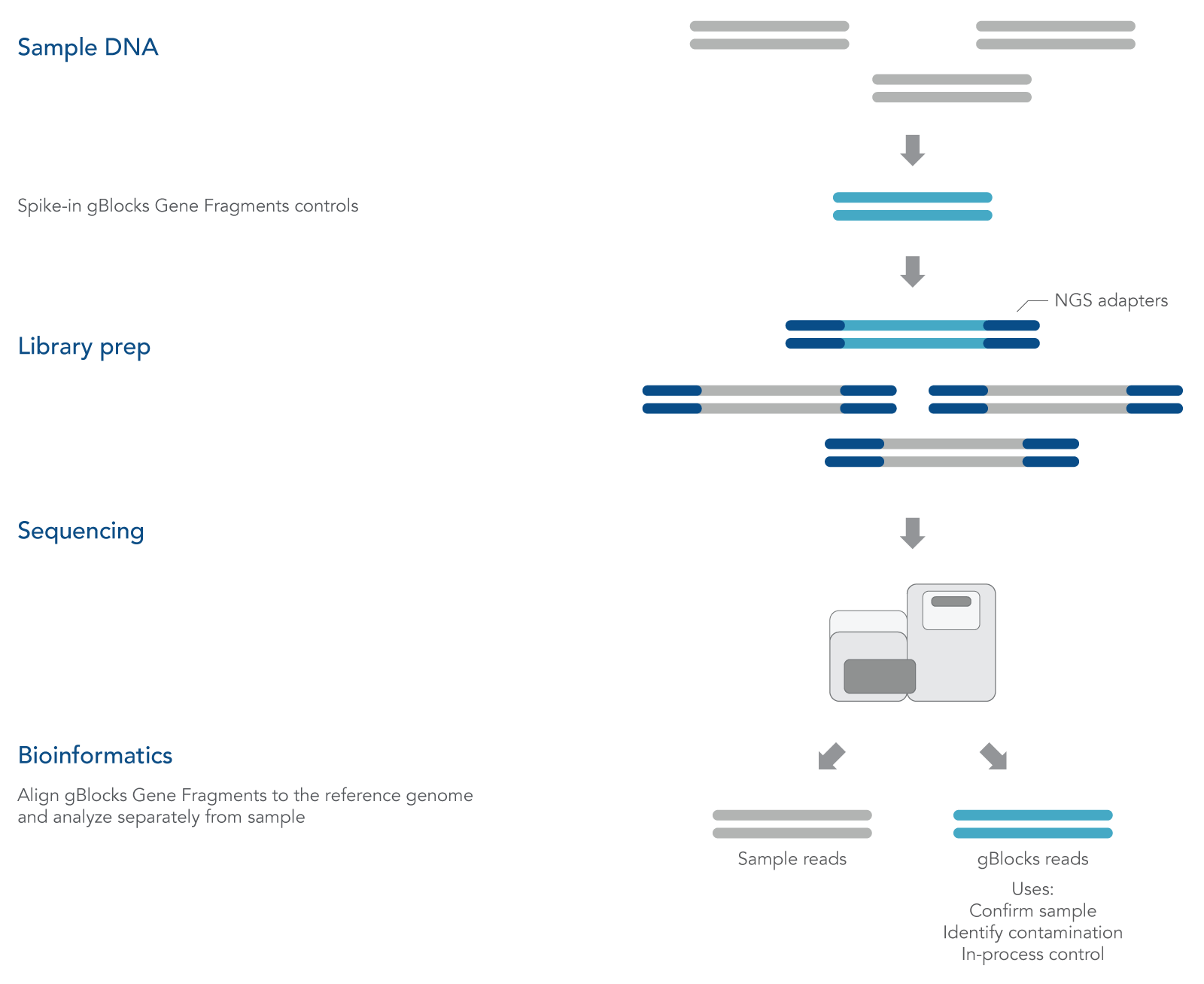
Sample tracking
Cross-contamination or sample swaps during processing and library prep could result in misassigned reads [2, 3]. To mitigate this risk, a unique gBlocks Gene Fragments control fragment can be spiked into each sample and used to track the presence of its associated target DNA sequence during data analysis to readily identify any sample swap events. This approach can help identify any sample swap events.
Controls for sample tracking are typically added to samples either before DNA fragmentation (so that the gBlocks Gene Fragments control fragment is subject to the same fragmentation conditions as the target DNA) or added after fragmentation if the gBlocks Gene Fragments lengths are similar to the mean fragment size.
Design recommendations:
- gBlocks Gene Fragments controls should bear no homology to the target sequence.
- When using multiple gBlocks Gene Fragments controls to track samples, make sure that there is no homology between the control sequences.
- Fragmentation technique (enzymatic or mechanical) should be considered when designing a gBlocks Gene Fragments control if spiking in before fragmentation. Complexities within sequences such as GC extremes could influence enzymatic fragmentation.
- If samples are derived from cell-free DNA, gBlocks Gene Fragments controls can be designed to match target DNA lengths (125–250 bp) without the need for fragmentation.
- For targeted sequencing using hybridization capture, a probe designed against the gBlocks Gene Fragments control sequences should be spiked into the panel.
SNP detection in amplicon sequencing
Sample tracking can also be used when detecting single nucleotide polymorphisms (SNPs) by amplicon sequencing techniques, such as the IDT xGen™ Amplicon Technology found in our predesigned amplicon panels or custom panels. With these applications, gBlocks Gene Fragments controls can be designed to encode the region flanking the SNP, while a known barcode or mutant sequence can be incorporated in place of the SNP as a marker. This barcode or mutant sequence differentiates control reads from those derived from experimental reads, making sample tracking more efficient.
Design recommendations:
- Primer binding sites for amplifying the SNP region can be represented in the gBlocks Gene Fragments so that the same set of primers is used to amplify both the target and the control.
- The length and GC profile of gBlocks Gene Fragments controls should resemble those of the target amplicon sequences.
Hybridization capture
For hybridization capture-based targeted sequencing approaches, such as those employing xGen Predesigned Hyb Panels or xGen Custom Hyb Panels, gBlocks Gene Fragments controls can be used to test capture efficiency and detect if there is any bias in the capture. These synthetic controls with the sequence of interest can be spiked into samples before library prep at known concentrations (or copy numbers) and quantified post-capture by qPCR to measure relative capture efficiencies for target vs. control sequences. Controls can also be designed to represent different experimental conditions (e.g., a range of GC content: 20%, 40%, 60%, and 80%) and measure capture efficiencies within the designated range.
Design recommendations:
- If the control is sufficiently homologous to the target sequence with only a few mutations, then a separate probe against the control would not need to be spiked into the panel/probe pool.
- If the controls represent different experimental conditions and do not resemble the target sequence, then probes complementary to the controls must be spiked into the panel/probe pool.
- The gBlocks Gene Fragments controls and complementary probes spiked into the sample and panel respectively should not affect the capture of the target sequences.
- Make sure that the relative copy numbers of the controls and their complementary probes are in the same range as the targets and their complementary probes.
Amplicon sequencing
Controls used in amplicon sequencing can indicate differences in amplification rates across targets of different lengths and GC content. To determine if there is amplification bias, gBlocks Gene Fragments are added at a known copy number and amplified simultaneously with the target sequence. Controls can also be quantified to normalize inputs across multiple targets before sequencing.
Design recommendations:
- The gBlocks Gene Fragments control sequence should bear no homology to the target amplicon sequence.
- Primer binding sites in the target sequence should be included in the associated gBlocks Gene Fragments sequence.
- Control sequences should be about the same length and GC content as target amplicon sequences.
Library preparation
For library preparation that includes PCR amplification, controls are also used to quickly check the rate of amplification errors that may be introduced during this step. Errors or mismatches in gBlocks Gene Fragments control reads of a known sequence can be compared to potential errors introduced in target reads. Here, the control sequence should not be homologous to the target, but it should be representative in length and GC content.
Controls can also help assess adapter ligation efficiencies by qPCR before sequencing. The gBlocks Gene Fragments controls can be designed to be in the same size range as the mean fragment size of the target DNA post-fragmentation. Control fragments can be spiked into the fragmented target DNA sample before the end-repair and A-tailing steps of library prep, for use with TA-ligation adapters such as TruSeq™-Compatible Full-length Adapters.
From platform adapters to our full portfolio of xGen products for targeted sequencing, IDT delivers innovative, high-quality NGS products to enable you to flex your discovery power. Consider using gBlocks Gene Fragments for sample tracking, quality checks, or as controls to test different experimental conditions in your next NGS assay. Learn more about customizing the design of gBlocks Gene Fragments by visiting this product page. For more information on resuspension, quantification, and copy number calculations of gBlocks Gene Fragments, see Tips for working with gBlocks Gene Fragments.

