What is xGen Adaptase technology?
xGen Adaptase technology is a highly efficient, template-independent adapter ligation reagent for single stranded DNA (ssDNA) substrates. Adaptase technology is suitable for NGS libraries constructed using denatured, bisulfite-converted DNA for methylation sequencing (methyl-seq), heavily damaged DNA samples where dsDNA prep is not suitable, metagenomic/virome samples comprising a mixture of DNA and dsDNA genomes, first strand cDNA samples following reverse transcription for RNA-seq and for DNA from single cells. Adaptase technology is used during NGS library preparation to add a stubby Read 2 adapter to the 3’ ends of ssDNA (Figure 1).
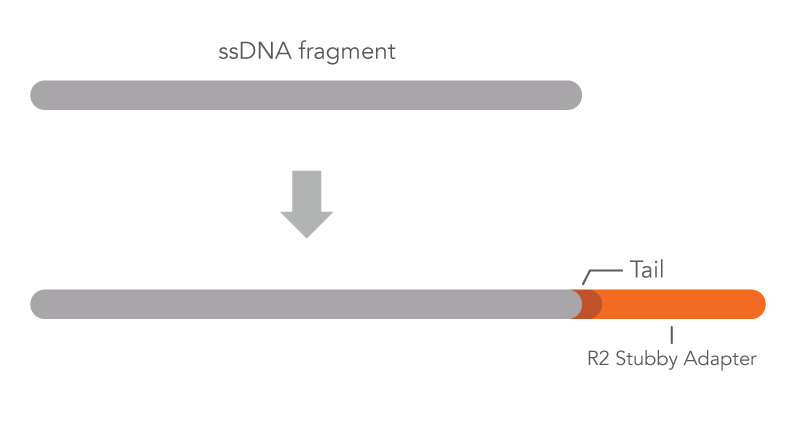
What you need to know about xGen Adaptase technology
- Functionality: During an NGS library prep workflow, xGen Adaptase technology is used to ligate adapters to the 3’ ends of ssDNA fragments with high efficiency.
-
Benefits:
- Produces directional libraries.
- Enables high-complexity libraries from bisulfite converted DNA by constructing the library post-conversion using ssDNA fragments. Traditional methods convert library preparation, which leads to significant loss of library complexity due to fragmentation of library molecules.
- Eliminates the polishing step in methyl-seq library prep workflows so the methylation status of 3’ ends of each fragment is preserved. This is advantageous because filling-in of 5’ overhangs by a polymerase during the polishing step introduces artificial cytosine content of up to 25 bp in length which requires trimming for accurate downstream analysis.
- Enables faster RNA-seq library prep as conventional second strand cDNA synthesis is not required.
- Uniquely maintains relative abundance of both ssDNA and dsDNA genomes from metagenomic and viromic samples.
- Supports low nanogram and picogram inputs since adapter titration is not required, maintaining library complexity at low input quantities.
- Compatibility: xGen Adaptase technology can be used with a variety of sample types including damaged samples that are heavily nicked, denatured, or heat attenuated since intact dsDNA is not required. It can also be used with metagenomic samples that require denaturing extraction methods to avoid representation bias from difficult-to-extract species. The xGen Methyl-Seq Library Prep Kit is also compatible with enzymatic conversion modules for methylation sequencing, and when used upstream of library prep, it overcomes the requirement for methylated adapters.
-
Applications:
- Methylation sequencing, including single cell (also referred to as bisulfite sequencing)
- RNA-seq
- Metagenomics
- Analysis of heavily damaged samples
- Research areas: xGen Adaptase technology can be incorporated into an NGS library prep workflow aimed at a variety of research areas from epigenetics to cancer research.
- Protocols: Download these kit protocols to see how xGen Adaptase technology is integrated into a variety of NGS library prep workflows:
Already using an Adaptase-based xGen Library Prep kit? Download the Tail trimming for better data technical note for details regarding bioinformatic recommendations when using xGen Adaptase technology.
What is xGen Normalase technology?
xGen Normalase technology can be used in NGS library prep workflows to complete enzymatic library normalization (Figure 2). Normalase technology streamlines normalization steps, by replacing individual library quantification and variable sample volume adjustments with equal volume pooling from each sample.
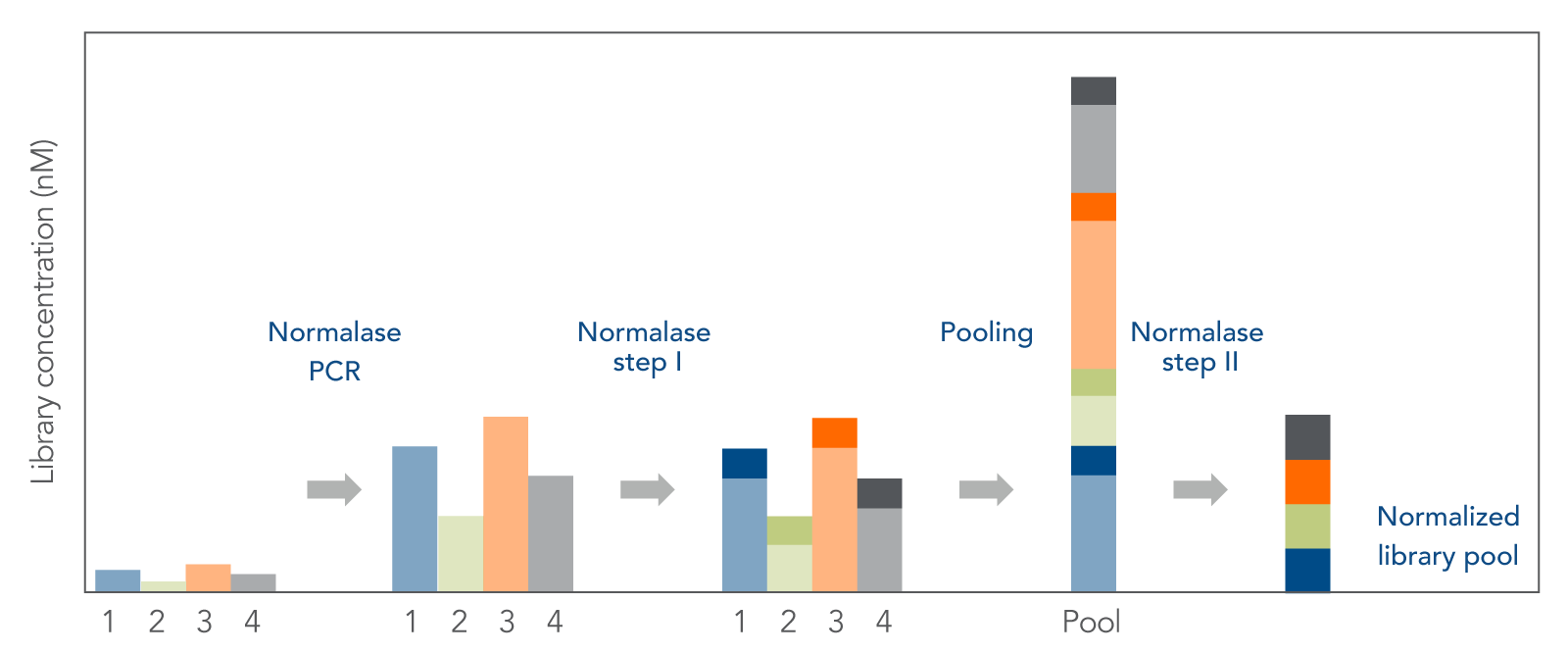
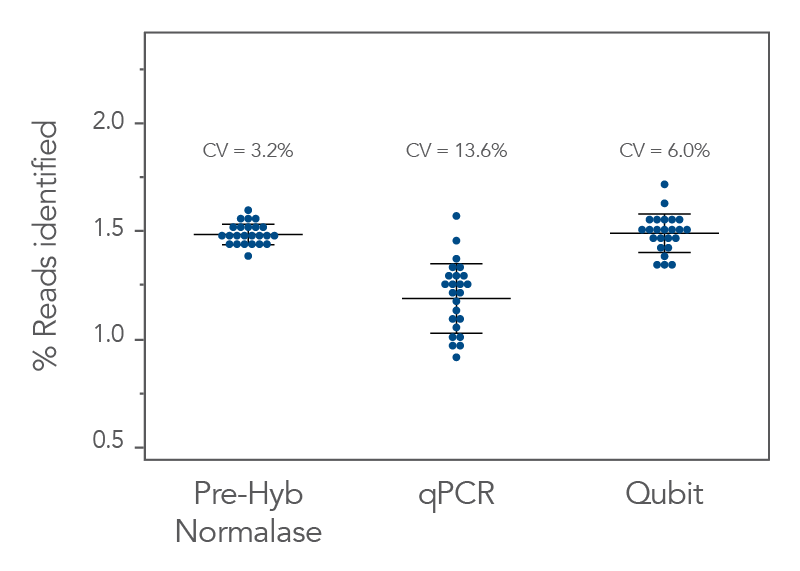
It also reduces sequencing costs by producing more even multiplexed sequencing within a pool. For the libraries represented below which were tested using the xGen Pre-Hybridization Capture Normalase Module (Figure 3) the coefficient of variation (CV) for a Normalase pool is <10%, which is more uniform than when normalizing library pools by qPCR or fluorometric methods.
What you need to know about xGen Normalase technology
- Functionality: xGen Normalase technology can be integrated into an NGS library prep workflow in which enzymatic library normalization is used. The Normalase workflow consists of a 15-minute incubation of condition library fragments (adapters have been ligated, etc); an equal volume pooling, and another 15-minute incubation.
-
Benefits:
- Saves time by cutting out individual library quantification and concentration adjustment steps after library prep.
- Generates better coefficient of variation (CV) values when compared to conventional normalization methods —i.e., qPCR and Qubit (Figure 3).
- Allows for more even multiplexing per run which reduces sequencing cost.
- Compatibility: xGen Normalase technology is compatible with library prep workflows that add index sequences via adapter ligation with full-length, indexed adapters, as well as workflows that add index sequences via PCR with stubby adapters. xGen Normalase technology should be used with libraries that have an amplified yield that meets the specified minimum threshold for library yield.
- Applications: xGen Normalase technology can be used in various sequencing applications through one of two normalization modules. The xGen Normalase Module can be used for direct sequencing applications such as whole genome sequencing (WGS) and whole transcriptome sequencing, while the xGen Pre-Hybridization Capture Normalase Module is ideal to use in targeted NGS library prep workflows before a hybridization capture. It is worth noting that the xGen Normalase technology cannot be used with PCR-free sequencing applications.
- Research areas: Since xGen Normalase technology can be easily incorporated into various NGS library prep workflows, the research areas that this technology can be applied to are numerous —i.e., anywhere a multiplexed, direct sequencing approach is used xGen Normalase technology can also be used. Similarly, for any application where multiplexed hybridization capture is being used, xGen Pre-Hybridization Capture Normalase technology can be used.
- Protocols: Download the xGen Normalase Module Protocol and the xGen Pre-Hybridization Capture Normalase Module Protocol to learn more about how Normalase technology can streamline your NGS library prep.
Curious about how well xGen Normalase technology integrates with an NGS library prep kit? Download the xGen Normalase Module application note.
RUO24-2752_001

