Oligonucleotide synthesis is a complex process that requires more than one hundred sequential chemical reactions to make a single, 25-base sequence. Contemporary synthesis chemistry is proven, and today's synthesis platforms are reliable and highly automated. Still, each oligonucleotide* synthesized at IDT is evaluated for quality before shipping to make sure that the correct sequence was made. One method to assess compound identity in a high throughput environment is mass spectrometry (MS).
What is mass spectrometry?
In MS analysis, a small amount of a synthesized oligonucleotide is ionized, then the ions are propelled into a mass detector/analyzer where molecular weight is measured. The analysis compares the calculated molecular weight of the given sequence to the measured molecular weight.
Mass spectrometry methods
Two methods of mass spectrometry are routinely used to analyze the mass of an oligonucleotide.
- MALDI-TOF (matrix-assisted laser desorption ionization-time of flight)
- ESI-MS (electrospray ionization mass spectrometry)
IDT currently uses ESI-MS as it maintains mass accuracy, resolution, and sensitivity for a molecular weight range that is more applicable to the product lengths IDT manufactures as compared to MALDI-TOF methods.
How to interpret mass spectrometry results
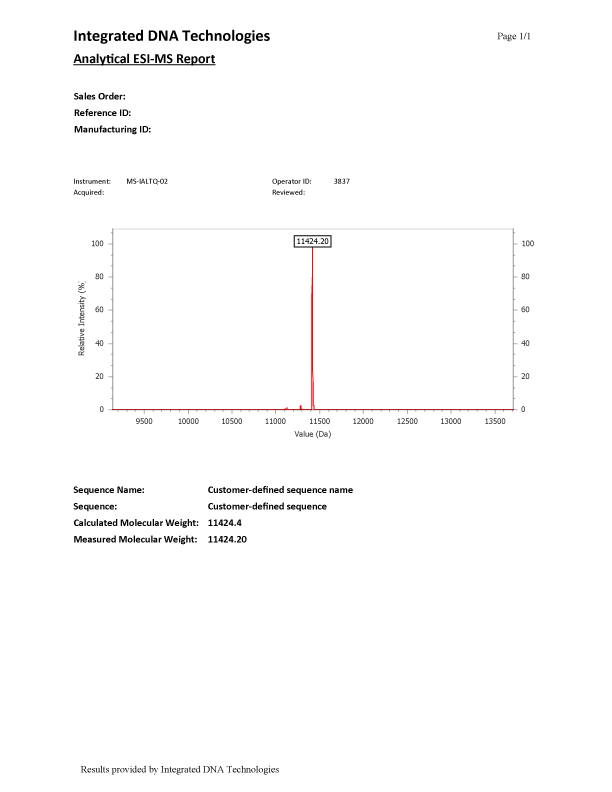
ESI-MS (Figure 1) results have a main peak representing the synthesized oligonucleotide, which is the labeled peak in the spectrum. Small peaks found in the spectrum in addition to the labeled full-length-product peak are normal and are discussed in greater detail below.
Additional peaks
There are various reasons why an ESI-MS report could show additional peaks. One common reason for secondary peaks in an ESI-MS are N-mers. N-mers (i.e., N-1, N-2, etc.) refer to deletions or truncations that occurred during the synthesis of the oligo and are detectable by MS. Deletions, commonly referred to as N-1, range from approximately 304–329 mass units less than the major peak, depending on the DNA base that is missing. Truncations, (N-2 or higher), will have varying mass differences less than the major peak.
Depurination can also occur during ESI analysis because of heating in the transport region of the ESI instrument and due to actual depurination of the sample before analysis. Depurination can create secondary peaks having approximately 135 (dA) or 151 (dG) mass units less than the major peak. Frequently, these species are created by the process as the sample is measured but are not present in the product itself.
In addition, synthetic oligonucleotides made using phosphoramidite chemistries employ protecting groups on the primary amines in dA, dC, and dG phosphoramidites to prevent branching and other undesired side reactions during chain elongation. Protecting groups are cleaved off during the final steps. Incomplete removal of these side groups results in peaks that have masses heavier than the major peak in the MS spectrum.
Finally, oligonucleotide modifications add mass to the product. Modifications commonly used in oligonucleotides are well characterized and their masses are considered in the final mass spectrograms produced. If a modification has not coupled fully, a peak less than the major peak may show in the MS spectrum corresponding to that modification's mass. A list of mass contributions for the most often requested modifications of DNA and RNA oligonucleotides are listed on IDT's Oligo modifications page. Also, if needed, the anhydrous molecular weight of both unmodified and modified oligonucleotides can be calculated using our OligoAnalyzer™ Tool.
It is important to note that IDT has internal standards for the amount of different truncations or adducts that can be found in an oligo and will remake oligonucleotides with a significant amount of additional peaks based on sequence type.
MS data is provided online for IDT orders
IDT offers ESI-MS QC free-of-charge on all standard oligonucleotides*. Find this QC data online for oligonucleotides that were ordered on the website once they have shipped. To view the data, follow these steps:
- Go to My Account > Order History and search for your order.
- Click the zip file located under the QC/COA heading for your order. This file contains all of the trace files in the order.
To get help with locating a trace, contact us.
*With the exception of mixed base oligos, which could potentially represent multiple sequences therefore cannot accurately be evaluated by ESI mass spectrometry
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical
diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. RUO22-1653_001